Simulation for Brain Networks
Extreme Scale Analytics and Simulation for Brain Networks
This workpackage is meant to enable the reconstruction of the human connectome, i.e. entire set of nerve fiber tracts that are resolvable down to the micrometer scale. This requires the serial application of two imaging methods, Diffusion MRI (dMRI) and 3D Polarized Light Imaging (3D-PLI), to the same brain tissue.
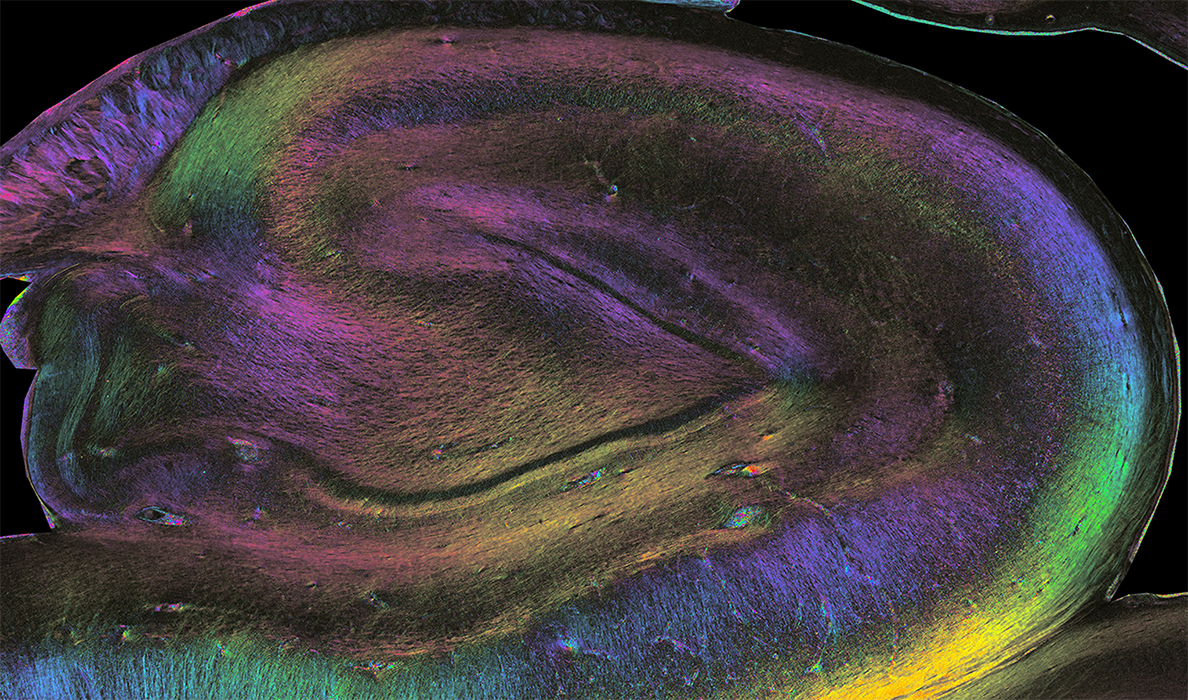
Color-encoded direction map stemming from a coronal slice of a human hippocampus scanned using Polarized Light Microscopy at the microscale” (credit: Pr Markus Axer, Fiber Architecture Group INM1, FZJ)”
The acquired data sets to be analyzed are of TByte (dMRI) and PByte (3D-PLI) size, respectively. Both imaging techniques provide orientation information, i.e. vectors or distribution functions, indicating the local orientations of fiber tracts. Such orientation information can be used to follow and model fiber tracts throughout the brain by means of a so-called tractography algorithm.
We have identified a global spin glass tractography algorithm to be the most promising and reliable approach available to date. However, it is also most computationally intensive, in particular when applied to PByte of data. This makes the efficient use of extreme scale machines indispensable, which is a fundamental goal of this workpackage. ExaTract is the spin glass-based tractography software we are currently pushing towards exascale, enabling us to reconstruct the human brain’s connectome at unprecedented detail.
Since the reliability of the used local orientation information is essential for the reliability of the tractography results (i.e. tractograms), we are also seeking to optimize the signal interpretation of the respective techniques by means of tissue modeling and AI-based measurement simulations.We have implemented the MEDUSA software to generate realistic numerical models of brain tissue (comprising neurons, astrocytes, and vessels), which can be input into the virtual MRI scanners or PLI microscopes to analyze the measurements according to the orientation of fiber tracts. Such models are useful to simulate the water diffusion process and AI-predict the expected MRI signal, for example
Team for Simulation for Brain Networks
Markus Axer
Team Leader
Markus Axer
Katrin Amunts
Co-Leader
Katrin Amunts
Cyril Poupon
Team Leader
Cyril Poupon
Jean-François Mangin
Co-Leader
Jean-François Mangin
Alexis Brullé
PhD
Alexis Brullé
Anas Bachiri
PhD
Anas Bachiri
Felix Matuschke
Postdoc
Felix Matuschke
Ivy Uszynski
Ivy Uszynski
Simon Legeay
PhD
